AR6 SLP Model Methodology¶
This page documents the methodology used to produce the AR6 sea level projection sub-models from the IPCC data. It is included because I'm not entirely sure what I did is statistically valid, so I thought it would be a good idea to document the process so any flaws I introduced can be pointed out.
import sys
import pandas as pd
import numpy as np
import plotly.graph_objects as go
from copy import deepcopy
from loguru import logger as l
from scipy import integrate
from scipy.stats import skewnorm, kstest
from numpy.typing import ArrayLike
from sunkcosts.model.ar6 import SLP
from sunkcosts.plotly import MINIMAL
_ = l.remove()
_ = l.add(
sys.stdout, colorize=True, format="[<green>{level}</green>] <cyan>{message}</cyan>"
)
First, I'll quickly note that the original data was in a NetCDF format that was a pain to work with. Accordingly, I built a dataclass for working with the data, which we'll use here.
More information on the data conversion process, and the .ssp (ie: JSON) files that the dataclass fetches the loaded data from can be found in this repository.
We'll start by loading the medium confidence SLP 245 scenario.
slp = SLP.load(confidence="medium", scenario="245")
We can view the scenario data as a dataframe.
slp.data.table
| 0.000 | 0.001 | 0.005 | 0.010 | 0.020 | 0.030 | 0.040 | 0.050 | 0.060 | 0.070 | ... | 0.930 | 0.940 | 0.950 | 0.960 | 0.970 | 0.980 | 0.990 | 0.995 | 0.999 | 1.000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2020 | 28 | 31 | 34 | 35 | 36 | 37 | 38 | 39 | 39 | 40 | ... | 70 | 72 | 73 | 76 | 79 | 85 | 97 | 112 | 138 | 165 |
| 2030 | 44 | 57 | 62 | 65 | 67 | 69 | 71 | 72 | 73 | 74 | ... | 134 | 138 | 141 | 146 | 153 | 163 | 187 | 213 | 267 | 318 |
| 2040 | 55 | 84 | 92 | 96 | 101 | 104 | 107 | 108 | 110 | 111 | ... | 207 | 213 | 219 | 227 | 238 | 255 | 296 | 336 | 408 | 490 |
| 2050 | 107 | 121 | 134 | 140 | 147 | 151 | 154 | 157 | 159 | 161 | ... | 303 | 310 | 318 | 330 | 346 | 374 | 428 | 492 | 589 | 714 |
| 2060 | 80 | 148 | 166 | 174 | 183 | 189 | 193 | 197 | 200 | 202 | ... | 397 | 407 | 419 | 434 | 457 | 497 | 570 | 646 | 789 | 947 |
| 2070 | 83 | 156 | 200 | 212 | 224 | 230 | 237 | 242 | 245 | 249 | ... | 509 | 521 | 537 | 556 | 587 | 638 | 722 | 825 | 1005 | 1207 |
| 2080 | 75 | 175 | 220 | 243 | 262 | 273 | 280 | 287 | 291 | 295 | ... | 627 | 643 | 663 | 690 | 730 | 789 | 904 | 1015 | 1244 | 1544 |
| 2090 | 84 | 177 | 256 | 273 | 295 | 308 | 318 | 326 | 333 | 340 | ... | 753 | 771 | 795 | 827 | 875 | 959 | 1092 | 1230 | 1509 | 1907 |
| 2100 | 106 | 175 | 277 | 305 | 332 | 351 | 362 | 371 | 380 | 387 | ... | 888 | 917 | 946 | 982 | 1038 | 1126 | 1283 | 1444 | 1777 | 2252 |
| 2110 | 207 | 285 | 324 | 342 | 366 | 379 | 391 | 400 | 409 | 416 | ... | 1030 | 1055 | 1086 | 1140 | 1205 | 1300 | 1496 | 1684 | 2035 | 2618 |
| 2120 | 224 | 311 | 356 | 376 | 401 | 418 | 431 | 442 | 452 | 459 | ... | 1161 | 1189 | 1227 | 1285 | 1359 | 1469 | 1691 | 1896 | 2303 | 2984 |
| 2130 | 238 | 335 | 386 | 408 | 436 | 455 | 470 | 482 | 493 | 502 | ... | 1293 | 1322 | 1367 | 1428 | 1513 | 1638 | 1885 | 2104 | 2569 | 3345 |
| 2140 | 254 | 357 | 415 | 440 | 469 | 492 | 508 | 522 | 534 | 543 | ... | 1424 | 1456 | 1506 | 1572 | 1668 | 1803 | 2079 | 2315 | 2833 | 3707 |
| 2150 | 269 | 378 | 443 | 470 | 502 | 528 | 545 | 560 | 572 | 584 | ... | 1552 | 1590 | 1643 | 1713 | 1817 | 1970 | 2269 | 2518 | 3089 | 4068 |
14 rows × 107 columns
A quick explanation of what this dataset is:
- Each row represents the sea level projections for a year.
- Each column represents a probability quantile.
- Each cell represents a sea level change in millimeters.
Sea Level Change
The sea level change is measured since the AR6 reference period, a quote is included below from the FACTS_confidence_output_file_readme that can be found in the original dataset.
The level of sea-level change since the AR6 reference period.
Honestly, I'm not sure what the AR6 reference period was exactly, so I kind of overlooked that. That is definitely a glaring limitation of this model/analysis.
Correction, it is:
relative to a baseline of 1995–2014
From: IPCC AR6 report, page 1302, table 9.9 commentary
Probability Quantile
I'm not entirely sure I understood the quantiles correctly. The wikipedia page for quantiles states that:
There is one fewer quantile than the number of groups created.
But this is not the case in this dataset, where there are 107 quantiles and 107 sea level change values.
The aforementioned FACTS_readme PDF says the following, but I still wasn't sure.
Quantiles of the probability box of the sea-level change variable. 107 quantiles are available, including 0.0 to 1.0 in 0.01 increments and a few key quantile values (i.e. 0.995, 0.999, etc.). Multiply the quantiles by 100 to get the equivalent percentile values.
What was initially unclear to me was whether the quantiles are the probability that a sea level change will be manifested, by a year, or they are implicitly probability buckets, or something else. At any rate, I asked ChatGPT and got the following answer:
In statistics, a quantile is a location in a set of data that divides the data into groups with the same number of values below and above it. Quantiles at 0 represent the minimum value in the dataset, while quantiles at 1 represent the maximum value. If this dataset is describing sea level changes, each quantile value would correspond to a different scenario of sea level change. In your DataFrame, the numbers along the top (0.000, 0.001, 0.005...etc.) are the quantiles. The numbers in the rows are the sea-level changes for those quantiles in that particular year. So, for example, the value of the cell in the 2030 row at the 0.001 quantile is 40 millimeters. This means that, in the 2030 scenario, there is a 0.1% probability that the sea level will rise 40 millimeters or less. Here's another example. In 2040, the value in the 0.030 quantile is 93 millimeters. This means that, in the 2040 scenario, there is a 3% probability that the sea level will rise 93 millimeters or less. These elements are cumulative: a quantile of 0.030 means that 3% of the total outcomes fall below this value.
This seemed to more or less make sense, because if we graph the quantiles for a year the graph matches loosely what we would expect from this description.
The data from 2030 is graphed below.
data = slp.data.table.loc[2030].reset_index()
ycol, xcol = ["quantile", "mm"]
data.columns = [ycol, xcol]
quantiles = np.array(data["quantile"].tolist())
sealevel = np.array(data["mm"].tolist())
fig = go.Figure()
fig.add_trace(
go.Scatter(x=sealevel, y=quantiles, mode="lines+markers", name="lines+markers")
)
fig.update_layout(
title=f"{ycol} vs. {xcol}",
title_x=0.5,
template=MINIMAL,
xaxis_title=xcol,
yaxis_title=ycol,
)
fig.update_layout(title_x=0.5, height=600)
fig.show(renderer="notebook")
This was somewhat confusing to me, but I think the way to interpret it is to say that the quantile is the probability the sea level will rise less than a given value. So, for instance, the 0.1 quantile (10% probability) is 77 millimeters. So we could make the following statements:
- There is a 10% probability that the sea level change we will see by the year 2030 will be less than 77 millimeters.
- There is a 100% probability that the sea level change we will see by the year 2030 will be less than 318 millimeters.
Another way of making the same statements would be (? - again, I'm not 100% my interpretation is correct):
- There is a 90% probability that the sea level change we will see by the year 2030 will be more than 77 millimeters.
- There is a 0% probability that the sea level change will see by the year 2030 will be more than 318 millimeters.
This intuitively interpretation intuitively makes sense to me, but I am also not that confident in it.
At any rate, now we want to convert this into a probability density function. I think (?) we can treat the data we have so far, effectively as a CDF (cumulative distribution function). As I understand, we can convert the CDF to a PDF by taking the first derivative of the CDF. However, when we plot the PDF we get this mess.
# take first derivative with numpy.gradient
pdf_values = np.gradient(quantiles, sealevel)
# remove any negative values obtained due to numerical errors
pdf_values = np.clip(pdf_values, a_min=0, a_max=None)
fig = go.Figure()
fig.add_trace(
go.Scatter(x=sealevel, y=pdf_values, mode="lines+markers", name="lines+markers")
)
fig.update_layout(
title=f"probability density vs. {xcol}",
title_x=0.5,
template=MINIMAL,
xaxis_title=xcol,
yaxis_title=ycol,
)
fig.update_layout(title_x=0.5, height=600)
fig.show(renderer="notebook")
Sorting the quantiles in ascending order doesn't seem to help much.
# Sort the sea level values and the quantiles in ascending order
sort_order = np.argsort(sealevel)
sort_sealevel = sealevel[sort_order]
sort_quantiles = quantiles[sort_order]
# take first derivative with numpy.gradient
sort_pdf_values = np.gradient(sort_quantiles, sort_sealevel)
# remove any negative values obtained due to numerical errors
sort_pdf_values = np.clip(sort_pdf_values, a_min=0, a_max=None)
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=sort_sealevel, y=sort_pdf_values, mode="lines+markers", name="lines+markers"
)
)
fig.update_layout(
title=f"probability density vs. {xcol}",
title_x=0.5,
template=MINIMAL,
xaxis_title=xcol,
yaxis_title=ycol,
)
fig.update_layout(title_x=0.5, height=600)
fig.show(renderer="notebook")
As a quick tangent, I also noticed there is this weird disjunction at the 50% quantile across the various distributions. I have no idea what is causing it. It is more visible when you plot all the series.
I checked and I'm 90% confident it's not something relating to me incorrectly parsing the data, but it's really strange and seems like either a mistake in the original model or data output from the IPCC (which intuitively I find unlikely) or just some strange artifact of the model they used to project sea level.
fig = go.Figure()
ycol, xcol = ["quantile", "mm"]
for year in slp.data._json["years"]:
temp_data = slp.data.table.loc[year].reset_index()
temp_data.columns = [ycol, xcol]
temp_quantiles = np.array(temp_data["quantile"].tolist())
temp_sealevel = np.array(temp_data["mm"].tolist())
fig.add_trace(
go.Scatter(
x=temp_sealevel, y=temp_quantiles, mode="lines+markers", name=str(year)
)
)
fig.update_layout(
title=f"{ycol} vs. {xcol}",
title_x=0.5,
template=MINIMAL,
xaxis_title=xcol,
yaxis_title=ycol,
)
fig.update_xaxes(range=[0, 2000])
fig.update_layout(title_x=0.5, height=600)
fig.show(renderer="notebook")
Moving forward...
So, interestingly, all of this investigation was done on the backdrop of me having written code to do this over a year ago, which at the time seemed to work perfectly. As I was revisiting this project, however, I started to wonder how the hell why code worked, because it didn't even factor in the quantiles. I still don't know entirely why it works, but I decided to use it, because I checked the outputs against the official AR6 report estimates, and somehow this model produces outputs which match up with the officially quoted figures. We'll review this now.
The underlying assumption that I think (?) I used was that the probability density functions would fundamentally reflect a skewed normal distribution.
This seems reasonable - we would expect a rough normal distribution, but long tail for higher sea level changes. The choppy PDF plot produced by taking the derivative of the quantile CDF above seems to support this hypothesis, and it seems reasonable on the basis of common sense (?).
Here's the class I made to fit the skewed normal distribution. Notebly, we're only using the sea level changes distribution. AIU scipy is using MLE under the hood to fit the distribution.
class SkewedNormalDistribution:
def __init__(self) -> None:
# self.model = lambda x: x
self.a = None
self.loc = None
self.scale = None
self.__trained = False
def __check_fit(self) -> None:
if not self.__trained:
raise Exception("you must fit the model parameters first")
def model(self, x: int | float | ArrayLike) -> int | float | ArrayLike:
return skewnorm.pdf(x, self.a, self.loc, self.scale)
def fit(self, distribution: ArrayLike) -> None:
self.a, self.loc, self.scale = skewnorm.fit(distribution)
self.__trained = True
l.success(
f"distribution fit with params: a={self.a}, loc={self.loc}, scale={self.scale}"
)
@property
def vmean(self) -> float | int:
"mean x value of the distribution"
self.__check_fit()
return skewnorm.stats(self.a, self.loc, self.scale, moments="m") # type: ignore
@property
def pmean(self) -> float | int:
"probability density at mean value of the distribution"
self.__check_fit()
return self.model(self.vmean) # type: ignore
@property
def vmed(self) -> float | int:
"median x value of the distribution"
self.__check_fit()
return skewnorm.ppf(0.5, self.a, self.loc, self.scale) # type: ignore
@property
def pmed(self) -> float | int:
"probability density at median value of the distribution"
self.__check_fit()
return self.model(self.vmed) # type: ignore
@property
def vmax(self) -> float | int:
"x value at maximum probability value of PDF"
self.__check_fit()
return optimize.fmin(lambda x: -self.model(x), self.loc, disp=False)[0] # type: ignore
@property
def pmax(self) -> float | int:
"maximum probability value of PDF"
self.__check_fit()
result = optimize.fmin(lambda x: -self.model(x), self.loc, disp=False) # type: ignore
return self.model(result[0]) # type: ignore
@property
def pmid(self) -> float | int | ArrayLike:
"midpoint between meanp and maxp"
self.__check_fit()
return (self.pmean + self.pmax) / 2
@property
def vmid(self) -> float | int:
"midpoint between meanv and maxv"
self.__check_fit()
return (self.vmean + self.vmax) / 2
@property
def v_at_pmid(self) -> float | int:
self.__check_fit()
return optimize.fsolve(lambda x: self.model(x) - self.pmid, self.vmean)[0] # type: ignore
@property
def p_at_vmid(self) -> float | int | ArrayLike:
self.__check_fit()
return self.model(self.vmid)
def interval(
self,
lower: float = -float("inf"),
upper: float = float("inf"),
) -> float:
"integrated area under PDF curve for given lower and upper bound interval"
if not self.model is None:
return round(
integrate.quad(self.probability, lower, upper)[0],
4,
)
else:
raise Exception("you must fit the model parameters first")
def probability(self, x: float | ArrayLike) -> float | ArrayLike:
"PDF function value at input x"
if not self.model is None:
return self.model(x)
else:
raise Exception("you must fit the model parameters first")
Now I'll attempt to explain how this is working in a visual sense, while also documenting the assumptions I made.
Also, I'm switching to matplotlib for this part to simplify stuff for myself - ie: these visualizations aren't interactive.
import matplotlib.pyplot as plt
import numpy as np
plt.style.use("dark_background")
The first assumption is that the data follows a skewed normal distribution. If we take the distribution of sea level values (for the year of data we're looking at, 2030) and plot them on a histogram, we can see that this seems to be loosely reflected in the data.
It's important to note that the information about the quantiles is erased here. What does our distribution actually represent then? It is, when we fit a model to it (I think) essentially assuming that each of individual sea level values in the original data is a single random variable drawn from a distribution of equally weighted potential variables. Notably, this is not actually what the data is. The data is a set of sea level change values, which are coupled to quantiles. However, because the quantiles are very nearly evenly spaced, we aren't corrupting the underlying inference capacity too much by fitting a model distribution to it.
I'll try and clarify this.
I added a line to the same plot. If we consider the "area" under the "curve" (not yet fit) of the distribution on the left of the line, that area is effectively the preportion of values from the original data, wherein the sea level value was less than 100 millimeters.
So in a sense, we can (though this is not perfectly statistically valid, I think (?)) take the percentage of values less than 100 millimeters, and divide it by the total area under the curve, and say that that is the probability that by 2030 the sea level will have risen 100 millimeters or less.
plt.hist(sealevel, density=True, bins=30, alpha=0.5)
plt.xlabel("sea level mm")
plt.axvline(x=50, c="red")
plt.axvline(x=100, c="red")
plt.show()
We might be able to extend this as well - if we want the probability the sea level will be between 50 and 100 millimeters, we can (?) take the 'area' between the two lines shown above.
This area is abstract though, so it's helpful if we fit a distribution to it.
snd = SkewedNormalDistribution()
snd.fit(np.array(slp.data.table.loc[2030].values))
[SUCCESS] distribution fit with params: a=5.182231462895766, loc=66.65545648583796, scale=50.66653599973423
x = np.linspace(min(sealevel), max(sealevel), 100)
y = snd.probability(x)
# y = skewnorm.pdf(x, a=snd.a, loc=snd.loc+3, scale=snd.scale-14)
plt.hist(sealevel, density=True, bins=30, alpha=0.5)
plt.plot(x, y, "r")
plt.show()
Using the skewed normal distribution, we get a much smoother distribution, which allows us to analytically integrate certain areas under the curve to get a probability we will see that sea level change for whatever distribution we've fit the model on (in this case 2030).
Essentially, there are three major assumptions here:
- That we can ignore the quantile data and treat the sea level as a random variable.
- That this random variable is being selected from a skewed normal distribution, and each of the sea level value instances from our original distribution that we are fitting a curve to has an equal probability of appearing (the same value can appear multiple times, which is what increases the probability we would see that value, because it increases the area in that section of the distribution)
- The main issue being that the original data values for sea level change were quantile based, but we're treating them as random variables.
- That the curve we fit is accurate.
Assumption/issue (1) and (2) we can essentially "ignore" - under the strict assumption that the outputs our model provides match up with the outputs the IPCC provides in their report (we will review this momentarily).
Assumption/issue (3) we can test by validating the model with the Kolmogorov-Smirnov test for goodness-of-fit. We'll start with this.
The two main components of the kstest we want to focus on are:
- Statistic: The statistic field refers to the KS statistic - the maximum difference between the EDF and the CDF. It is contrained between 0 and 1. The closer this value to 0, the better the fit, as the observed data closely follows the fitted distribution.
- p-value: This provides the probability for the null hypothesis of the KS test, which is that the data follow the specified distribution. A smaller p-value suggests that you can reject the null hypothesis, i.e., it suggests that the data does not follow the fitted distribution. Smaller values are better, and a common value for statistical significance is 0.05.
results = dict()
for year in slp.data._json["years"]:
snd = SkewedNormalDistribution()
values = np.array(slp.data.table.loc[year].values)
snd.fit(values)
cdf = lambda x: skewnorm.cdf(x, a=snd.a, loc=snd.loc, scale=snd.scale)
test = kstest(values, cdf)
results[year] = {
"statistic": round(test.statistic, 4),
"pvalue": round(test.pvalue, 4),
}
[SUCCESS] distribution fit with params: a=5.743975740306972, loc=36.164234604470565, scale=25.648743180786962 [SUCCESS] distribution fit with params: a=5.182231462895766, loc=66.65545648583796, scale=50.66653599973423 [SUCCESS] distribution fit with params: a=4.826670666558719, loc=98.77597705691915, scale=80.71888977645303 [SUCCESS] distribution fit with params: a=6.097923283953435, loc=143.43576407889822, scale=117.52710654937184 [SUCCESS] distribution fit with params: a=4.604312301237858, loc=177.73431558284676, scale=161.17478717133943 [SUCCESS] distribution fit with params: a=4.302638376657248, loc=217.3774712332787, scale=211.59553546919736 [SUCCESS] distribution fit with params: a=4.188282356556575, loc=253.6808747205983, scale=272.53183761119806 [SUCCESS] distribution fit with params: a=4.250054124385981, loc=289.71291494458524, scale=338.9957902267304 [SUCCESS] distribution fit with params: a=4.281880127311123, loc=328.98451633078946, scale=406.535462481623 [SUCCESS] distribution fit with params: a=6.673061154188609, loc=350.57018541954324, scale=490.7650896090627 [SUCCESS] distribution fit with params: a=6.75855006960453, loc=384.72780117768656, scale=560.9933986515815 [SUCCESS] distribution fit with params: a=6.787120353791238, loc=417.6704656895899, scale=631.2479293880192 [SUCCESS] distribution fit with params: a=6.8403784471332525, loc=449.7551450384476, scale=701.49002417919 [SUCCESS] distribution fit with params: a=6.881719567282601, loc=480.77103780606205, scale=771.0365584054249
We can review the results below.
results
{2020: {'statistic': 0.149, 'pvalue': 0.0154},
2030: {'statistic': 0.1427, 'pvalue': 0.0231},
2040: {'statistic': 0.1394, 'pvalue': 0.0283},
2050: {'statistic': 0.1499, 'pvalue': 0.0145},
2060: {'statistic': 0.1481, 'pvalue': 0.0163},
2070: {'statistic': 0.1451, 'pvalue': 0.0198},
2080: {'statistic': 0.1393, 'pvalue': 0.0284},
2090: {'statistic': 0.1386, 'pvalue': 0.0297},
2100: {'statistic': 0.1325, 'pvalue': 0.0425},
2110: {'statistic': 0.1471, 'pvalue': 0.0174},
2120: {'statistic': 0.1482, 'pvalue': 0.0162},
2130: {'statistic': 0.1488, 'pvalue': 0.0156},
2140: {'statistic': 0.1495, 'pvalue': 0.015},
2150: {'statistic': 0.1496, 'pvalue': 0.0148}}
The skewed normal distribution models seem to fit fairly well to the data, but we haven't addressed the underlying assumption about representing quantiles as random variables yet, which is (I think?) not strictly speaking statistically valid.
I think that this isn't too important though, so long as the outputs from the models here actually match up with the projections given by the IPCC.
The SND model is integrated into the SLP class (part of the package utils created for this project), so we can use this. Calling the train method fits a skewed normal distribution to each of the SLC arrays for the particular scenario and confidence level we're looking at.
slp.forecast.train()
[SUCCESS] distribution fit with params: a=5.743975740306972, loc=36.164234604470565, scale=25.648743180786962 [SUCCESS] distribution fit with params: a=5.182231462895766, loc=66.65545648583796, scale=50.66653599973423 [SUCCESS] distribution fit with params: a=4.826670666558719, loc=98.77597705691915, scale=80.71888977645303 [SUCCESS] distribution fit with params: a=6.097923283953435, loc=143.43576407889822, scale=117.52710654937184 [SUCCESS] distribution fit with params: a=4.604312301237858, loc=177.73431558284676, scale=161.17478717133943 [SUCCESS] distribution fit with params: a=4.302638376657248, loc=217.3774712332787, scale=211.59553546919736 [SUCCESS] distribution fit with params: a=4.188282356556575, loc=253.6808747205983, scale=272.53183761119806 [SUCCESS] distribution fit with params: a=4.250054124385981, loc=289.71291494458524, scale=338.9957902267304 [SUCCESS] distribution fit with params: a=4.281880127311123, loc=328.98451633078946, scale=406.535462481623 [SUCCESS] distribution fit with params: a=6.673061154188609, loc=350.57018541954324, scale=490.7650896090627 [SUCCESS] distribution fit with params: a=6.75855006960453, loc=384.72780117768656, scale=560.9933986515815 [SUCCESS] distribution fit with params: a=6.787120353791238, loc=417.6704656895899, scale=631.2479293880192 [SUCCESS] distribution fit with params: a=6.8403784471332525, loc=449.7551450384476, scale=701.49002417919 [SUCCESS] distribution fit with params: a=6.881719567282601, loc=480.77103780606205, scale=771.0365584054249
Now we can compare the predictions of the SLP SND model fits to the official IPCC forecasts. Before we do that though, I'll take a quick aside to mention that some properties to find the maximum probability value of the PDF function and also the x value at the maximum probability value were also added.
There are also meanp, and meanv values, which relate more to the "center of gravity" of the distribution, and not the maximum.
If that was confusing, I've also included some visualizations to assist in intuitively understanding them.
Maximum
x = np.linspace(0, 300, 100)
y = slp.forecast.probability(x, 2030)
plt.title("PDF curve of global sea level change by 2030")
plt.plot(x, y)
plt.axvline(slp.forecast.vmax(2030), c="red")
plt.axhline(slp.forecast.pmax(2030), c="red")
plt.show()
print(f"2030 vmax: {slp.forecast.vmax(2030)}")
print(f"2030 pmax: {slp.forecast.pmax(2030)}")
Mean
x = np.linspace(0, 300, 100)
y = slp.forecast.probability(x, 2030)
plt.title("PDF curve of global sea level change by 2030")
plt.plot(x, y)
plt.axvline(slp.forecast.vmean(2030), c="red")
plt.axhline(slp.forecast.pmean(2030), c="red")
plt.show()
print(f"2030 vmean: {slp.forecast.vmean(2030)}")
print(f"2030 pmean: {slp.forecast.pmean(2030)}")
Alltogether they look like the following chart. Two more points are added: medv - which is the median value, and midv, which is the midpoint between the sea level change at the max probability of the PDF and the mean.
x = np.linspace(0, 300, 100)
y = slp.forecast.probability(x, 2030)
plt.title("PDF curve of global sea level change by 2030")
plt.plot(x, y)
plt.axvline(slp.forecast.vmax(2030), c="red", label="max", alpha=0.6)
plt.axhline(slp.forecast.pmax(2030), c="red", alpha=0.6)
plt.axvline(slp.forecast.vmean(2030), c="blue", label="mean", alpha=0.6)
plt.axhline(slp.forecast.pmean(2030), c="blue", alpha=0.6)
plt.axvline(slp.forecast.vmed(2030), c="green", label="median", alpha=0.6)
plt.axhline(slp.forecast.pmed(2030), c="green", alpha=0.6)
plt.axvline(slp.forecast.vmid(2030), c="orange", label="midpoint", alpha=0.6)
plt.axhline(slp.forecast.p_at_vmid(2030), c="orange", alpha=0.6)
plt.legend()
plt.xlim([80, 120])
plt.ylim([0.01, 0.015])
plt.show()
Next, we'll check to make sure the predictions actually match up with the expected outputs.
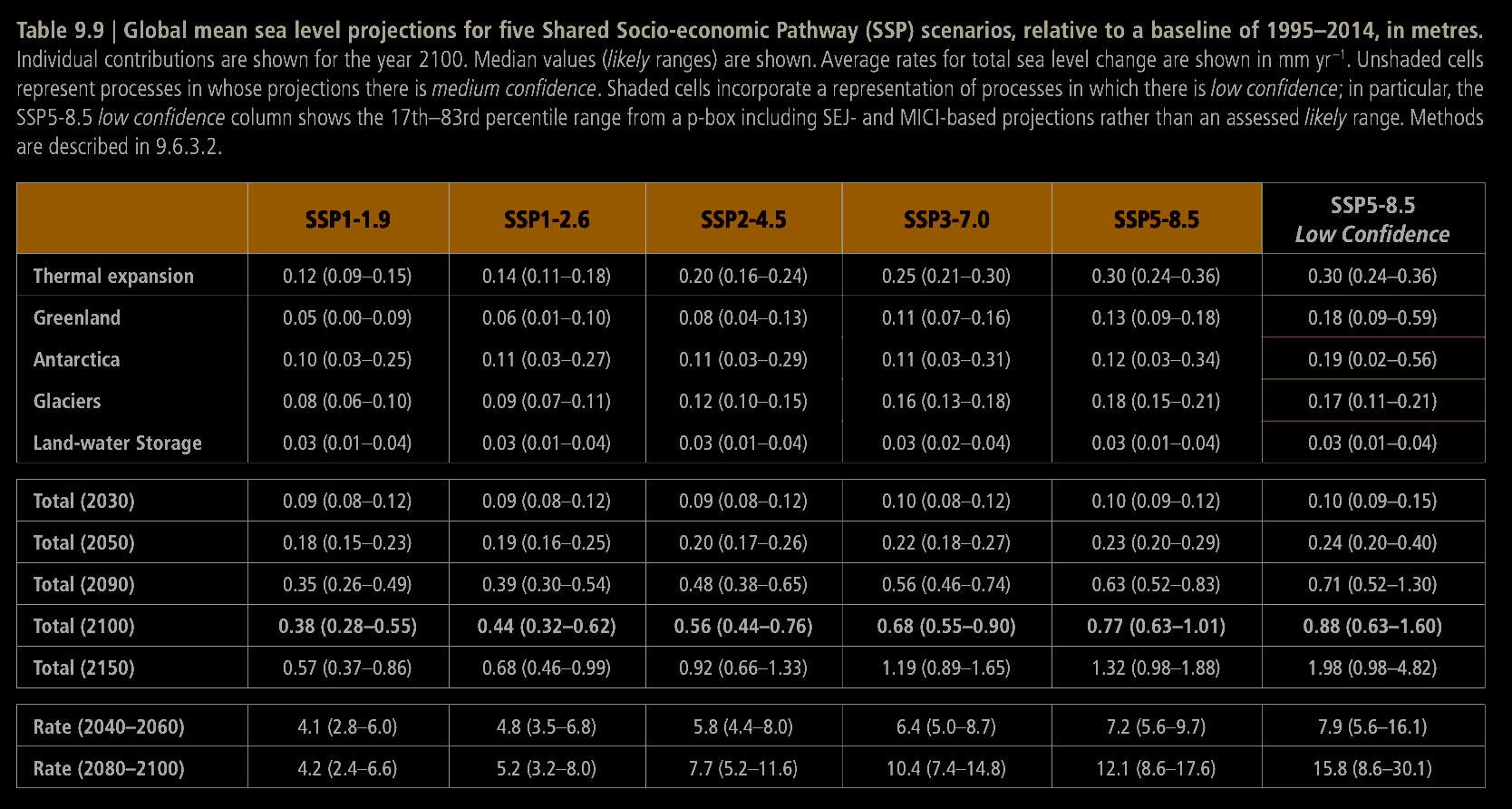
We'll use the following table, which is pulled from the IPCC AR6 report and can be found on page 1302 (I flipped the colors as I prefer dark mode).
Also, note that there are extra dashes and dots in the table columns for the different ssp scenarios. They are otherwise the same scenarios we're considering.
I manually extracted this data into the following table. I'm using the central values they provide, and ignoring the confidence interval. The objective here is just to tell whether the skewed normal fits match up loosely with the expected values. We'll also multiply the values in this dataframe by 1000 to account for the unit difference (our model outputs millimeters, they use meters).
years = [2030, 2050, 2090, 2100, 2150]
values = {
"126": [0.09, 0.19, 0.39, 0.44, 0.68],
"245": [0.09, 0.20, 0.48, 0.56, 0.92],
"585": [0.10, 0.23, 0.63, 0.77, 1.32],
}
val_df = pd.DataFrame(values, index=years)
val_df = val_df * 1000
val_df
| 126 | 245 | 585 | |
|---|---|---|---|
| 2030 | 90.0 | 90.0 | 100.0 |
| 2050 | 190.0 | 200.0 | 230.0 |
| 2090 | 390.0 | 480.0 | 630.0 |
| 2100 | 440.0 | 560.0 | 770.0 |
| 2150 | 680.0 | 920.0 | 1320.0 |
Now we can compare this against our models. We're focusing on the 'medium' confidence scenarios.
We'll try the median, which is what they use, but notably their median is different than ours. Actually I'm not even sure what their median is exactly referring to, but it probably factors in the quantiles somehow, which are ignored here.
scenarios = val_df.columns
diff_df = deepcopy(val_df)
comp_df = deepcopy(val_df)
for scen in scenarios:
slp = SLP.load(confidence="medium", scenario=scen)
slp.forecast.train()
pred = pd.Series([slp.forecast.vmed(year) for year in years], index=years)
comp_df[f"model_{scen}"] = pred
diff_df[scen] = (round((pred / diff_df[scen]) - 1, 4)) * 100
diff_df["avg_percent_diff"] = diff_df.mean(axis=1)
[SUCCESS] distribution fit with params: a=6.336008526116064, loc=36.217225776201914, scale=26.499101164646206 [SUCCESS] distribution fit with params: a=5.395765778925574, loc=65.96713161669996, scale=51.89790726109507 [SUCCESS] distribution fit with params: a=5.2516139267483695, loc=91.79716693057506, scale=84.3977925731794 [SUCCESS] distribution fit with params: a=5.375537285516717, loc=127.07699100029154, scale=120.50862925192759 [SUCCESS] distribution fit with params: a=4.75210367967559, loc=148.9815539003562, scale=161.9565510001559 [SUCCESS] distribution fit with params: a=4.9794339762155015, loc=181.0611547138783, scale=204.96800639226774 [SUCCESS] distribution fit with params: a=3.7235340713646448, loc=197.14713750731312, scale=259.8152957042531 [SUCCESS] distribution fit with params: a=3.8853618584306986, loc=216.11771757184601, scale=309.2036028951542 [SUCCESS] distribution fit with params: a=3.609209400576966, loc=230.41995592402873, scale=361.2054271926496 [SUCCESS] distribution fit with params: a=6.371515168495243, loc=254.63459928563884, scale=420.5578295282163 [SUCCESS] distribution fit with params: a=6.499318451859498, loc=271.9702126298085, scale=472.8110066298254 [SUCCESS] distribution fit with params: a=6.604161455986308, loc=287.0527075970766, scale=524.7800963533753 [SUCCESS] distribution fit with params: a=6.729952755482406, loc=301.1037168259759, scale=576.8664760591203 [SUCCESS] distribution fit with params: a=6.836750088876363, loc=314.06869887162077, scale=629.1149063926257 [SUCCESS] distribution fit with params: a=5.743975740306972, loc=36.164234604470565, scale=25.648743180786962 [SUCCESS] distribution fit with params: a=5.182231462895766, loc=66.65545648583796, scale=50.66653599973423 [SUCCESS] distribution fit with params: a=4.826670666558719, loc=98.77597705691915, scale=80.71888977645303 [SUCCESS] distribution fit with params: a=6.097923283953435, loc=143.43576407889822, scale=117.52710654937184 [SUCCESS] distribution fit with params: a=4.604312301237858, loc=177.73431558284676, scale=161.17478717133943 [SUCCESS] distribution fit with params: a=4.302638376657248, loc=217.3774712332787, scale=211.59553546919736 [SUCCESS] distribution fit with params: a=4.188282356556575, loc=253.6808747205983, scale=272.53183761119806 [SUCCESS] distribution fit with params: a=4.250054124385981, loc=289.71291494458524, scale=338.9957902267304 [SUCCESS] distribution fit with params: a=4.281880127311123, loc=328.98451633078946, scale=406.535462481623 [SUCCESS] distribution fit with params: a=6.673061154188609, loc=350.57018541954324, scale=490.7650896090627 [SUCCESS] distribution fit with params: a=6.75855006960453, loc=384.72780117768656, scale=560.9933986515815 [SUCCESS] distribution fit with params: a=6.787120353791238, loc=417.6704656895899, scale=631.2479293880192 [SUCCESS] distribution fit with params: a=6.8403784471332525, loc=449.7551450384476, scale=701.49002417919 [SUCCESS] distribution fit with params: a=6.881719567282601, loc=480.77103780606205, scale=771.0365584054249 [SUCCESS] distribution fit with params: a=6.35241015973173, loc=35.99658069718064, scale=28.111580396222273 [SUCCESS] distribution fit with params: a=5.096871305510005, loc=70.66973343458793, scale=54.96140440412516 [SUCCESS] distribution fit with params: a=5.763663587377232, loc=110.74874004286636, scale=89.83310367254231 [SUCCESS] distribution fit with params: a=5.228228021043594, loc=163.5534544090983, scale=133.38997146068937 [SUCCESS] distribution fit with params: a=5.101378964357277, loc=213.72709579057965, scale=186.94999434641414 [SUCCESS] distribution fit with params: a=5.636522073675502, loc=274.8963368738746, scale=251.0756454178623 [SUCCESS] distribution fit with params: a=4.782617220601494, loc=334.7672629463968, scale=331.61832359032053 [SUCCESS] distribution fit with params: a=5.308169643484584, loc=414.5584482139646, scale=416.1315973696705 [SUCCESS] distribution fit with params: a=5.22946285210722, loc=489.93843964346445, scale=523.6084467375629 [SUCCESS] distribution fit with params: a=6.757180784739575, loc=492.1731895549258, scale=658.6826221257295 [SUCCESS] distribution fit with params: a=6.732781229967694, loc=557.3925777094735, scale=762.216229241717 [SUCCESS] distribution fit with params: a=6.769984719450592, loc=619.7033640925753, scale=864.7711249649267 [SUCCESS] distribution fit with params: a=6.862736894866174, loc=677.0954078594514, scale=968.2556837673278 [SUCCESS] distribution fit with params: a=6.979824091070947, loc=729.4440840890079, scale=1071.8957799258405
comp_df
| 126 | 245 | 585 | model_126 | model_245 | model_585 | |
|---|---|---|---|---|---|---|
| 2030 | 90.0 | 90.0 | 100.0 | 100.971427 | 100.828952 | 107.739857 |
| 2050 | 190.0 | 200.0 | 230.0 | 208.358061 | 222.706512 | 253.522322 |
| 2090 | 390.0 | 480.0 | 630.0 | 424.568615 | 518.315754 | 695.231726 |
| 2100 | 440.0 | 560.0 | 770.0 | 473.816901 | 603.137238 | 843.101882 |
| 2150 | 680.0 | 920.0 | 1320.0 | 738.400219 | 1000.827256 | 1452.426764 |
diff_df
| 126 | 245 | 585 | avg_percent_diff | |
|---|---|---|---|---|
| 2030 | 12.19 | 12.03 | 7.74 | 10.653333 |
| 2050 | 9.66 | 11.35 | 10.23 | 10.413333 |
| 2090 | 8.86 | 7.98 | 10.35 | 9.063333 |
| 2100 | 7.69 | 7.70 | 9.49 | 8.293333 |
| 2150 | 8.59 | 8.79 | 10.03 | 9.136667 |
We can rerun this with the midpoint.
scenarios = val_df.columns
diff_df = deepcopy(val_df)
comp_df = deepcopy(val_df)
for scen in scenarios:
slp = SLP.load(confidence="medium", scenario=scen)
slp.forecast.train()
pred = pd.Series([slp.forecast.vmid(year) for year in years], index=years)
comp_df[f"model_{scen}"] = pred
diff_df[scen] = (round((pred / diff_df[scen]) - 1, 4)) * 100
diff_df["avg_percent_diff"] = diff_df.mean(axis=1)
[SUCCESS] distribution fit with params: a=6.336008526116064, loc=36.217225776201914, scale=26.499101164646206 [SUCCESS] distribution fit with params: a=5.395765778925574, loc=65.96713161669996, scale=51.89790726109507 [SUCCESS] distribution fit with params: a=5.2516139267483695, loc=91.79716693057506, scale=84.3977925731794 [SUCCESS] distribution fit with params: a=5.375537285516717, loc=127.07699100029154, scale=120.50862925192759 [SUCCESS] distribution fit with params: a=4.75210367967559, loc=148.9815539003562, scale=161.9565510001559 [SUCCESS] distribution fit with params: a=4.9794339762155015, loc=181.0611547138783, scale=204.96800639226774 [SUCCESS] distribution fit with params: a=3.7235340713646448, loc=197.14713750731312, scale=259.8152957042531 [SUCCESS] distribution fit with params: a=3.8853618584306986, loc=216.11771757184601, scale=309.2036028951542 [SUCCESS] distribution fit with params: a=3.609209400576966, loc=230.41995592402873, scale=361.2054271926496 [SUCCESS] distribution fit with params: a=6.371515168495243, loc=254.63459928563884, scale=420.5578295282163 [SUCCESS] distribution fit with params: a=6.499318451859498, loc=271.9702126298085, scale=472.8110066298254 [SUCCESS] distribution fit with params: a=6.604161455986308, loc=287.0527075970766, scale=524.7800963533753 [SUCCESS] distribution fit with params: a=6.729952755482406, loc=301.1037168259759, scale=576.8664760591203 [SUCCESS] distribution fit with params: a=6.836750088876363, loc=314.06869887162077, scale=629.1149063926257 [SUCCESS] distribution fit with params: a=5.743975740306972, loc=36.164234604470565, scale=25.648743180786962 [SUCCESS] distribution fit with params: a=5.182231462895766, loc=66.65545648583796, scale=50.66653599973423 [SUCCESS] distribution fit with params: a=4.826670666558719, loc=98.77597705691915, scale=80.71888977645303 [SUCCESS] distribution fit with params: a=6.097923283953435, loc=143.43576407889822, scale=117.52710654937184 [SUCCESS] distribution fit with params: a=4.604312301237858, loc=177.73431558284676, scale=161.17478717133943 [SUCCESS] distribution fit with params: a=4.302638376657248, loc=217.3774712332787, scale=211.59553546919736 [SUCCESS] distribution fit with params: a=4.188282356556575, loc=253.6808747205983, scale=272.53183761119806 [SUCCESS] distribution fit with params: a=4.250054124385981, loc=289.71291494458524, scale=338.9957902267304 [SUCCESS] distribution fit with params: a=4.281880127311123, loc=328.98451633078946, scale=406.535462481623 [SUCCESS] distribution fit with params: a=6.673061154188609, loc=350.57018541954324, scale=490.7650896090627 [SUCCESS] distribution fit with params: a=6.75855006960453, loc=384.72780117768656, scale=560.9933986515815 [SUCCESS] distribution fit with params: a=6.787120353791238, loc=417.6704656895899, scale=631.2479293880192 [SUCCESS] distribution fit with params: a=6.8403784471332525, loc=449.7551450384476, scale=701.49002417919 [SUCCESS] distribution fit with params: a=6.881719567282601, loc=480.77103780606205, scale=771.0365584054249 [SUCCESS] distribution fit with params: a=6.35241015973173, loc=35.99658069718064, scale=28.111580396222273 [SUCCESS] distribution fit with params: a=5.096871305510005, loc=70.66973343458793, scale=54.96140440412516 [SUCCESS] distribution fit with params: a=5.763663587377232, loc=110.74874004286636, scale=89.83310367254231 [SUCCESS] distribution fit with params: a=5.228228021043594, loc=163.5534544090983, scale=133.38997146068937 [SUCCESS] distribution fit with params: a=5.101378964357277, loc=213.72709579057965, scale=186.94999434641414 [SUCCESS] distribution fit with params: a=5.636522073675502, loc=274.8963368738746, scale=251.0756454178623 [SUCCESS] distribution fit with params: a=4.782617220601494, loc=334.7672629463968, scale=331.61832359032053 [SUCCESS] distribution fit with params: a=5.308169643484584, loc=414.5584482139646, scale=416.1315973696705 [SUCCESS] distribution fit with params: a=5.22946285210722, loc=489.93843964346445, scale=523.6084467375629 [SUCCESS] distribution fit with params: a=6.757180784739575, loc=492.1731895549258, scale=658.6826221257295 [SUCCESS] distribution fit with params: a=6.732781229967694, loc=557.3925777094735, scale=762.216229241717 [SUCCESS] distribution fit with params: a=6.769984719450592, loc=619.7033640925753, scale=864.7711249649267 [SUCCESS] distribution fit with params: a=6.862736894866174, loc=677.0954078594514, scale=968.2556837673278 [SUCCESS] distribution fit with params: a=6.979824091070947, loc=729.4440840890079, scale=1071.8957799258405
comp_df
| 126 | 245 | 585 | model_126 | model_245 | model_585 | |
|---|---|---|---|---|---|---|
| 2030 | 90.0 | 90.0 | 100.0 | 95.528640 | 95.699737 | 102.257837 |
| 2050 | 190.0 | 200.0 | 230.0 | 195.760647 | 209.078618 | 239.912564 |
| 2090 | 390.0 | 480.0 | 630.0 | 400.967678 | 489.904611 | 652.204921 |
| 2100 | 440.0 | 560.0 | 770.0 | 448.375494 | 568.807158 | 789.667120 |
| 2150 | 680.0 | 920.0 | 1320.0 | 658.970697 | 903.027626 | 1315.112348 |
diff_df
| 126 | 245 | 585 | avg_percent_diff | |
|---|---|---|---|---|
| 2030 | 6.14 | 6.33 | 2.26 | 4.910000 |
| 2050 | 3.03 | 4.54 | 4.31 | 3.960000 |
| 2090 | 2.81 | 2.06 | 3.52 | 2.796667 |
| 2100 | 1.90 | 1.57 | 2.55 | 2.006667 |
| 2150 | -3.09 | -1.84 | -0.37 | -1.766667 |
The midpoint seems to work much better at matching the figures they provide. Either way, the skewed normal distribution models fit to just the sea level change data (represented as a distribution and ignoring quantiles) for each decade pretty well. Going from this fairly non-rigorous and unscientific eyeballing of the model outputs and the IPCC figures, they are usable, with somewhere maybe in the range of 5-10% error, acceptable for this project.